Measuring the selective packaging of RNA molecules by viral coat proteins in cells
Measuring the selective packaging of RNA molecules by viral coat proteins in cells
Rastandeh, A.; Makasarashvili, N.; Dhaliwal, H. K.; Subramanian, S.; Villarreal, D. A.; Baker, S.; Gamez, E. I.; Parent, K. N.; Garmann, R. F.
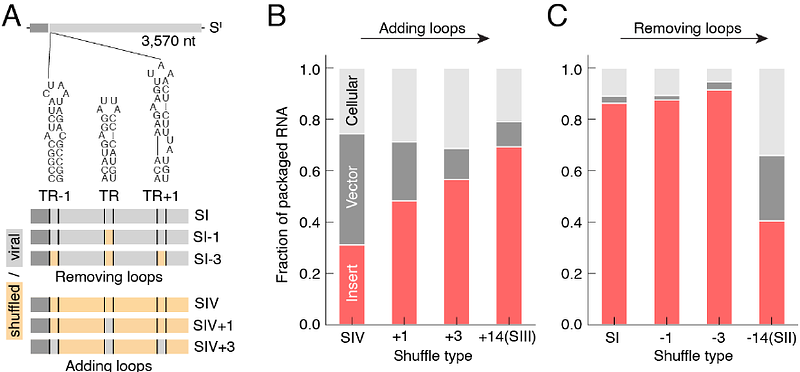
AbstractSome RNA viruses package their genomes with extraordinary selectivity, assembling protein capsids around their own viral RNA while excluding nearly all host RNA. How the assembling proteins distinguish viral RNA from host RNA is not fully understood, but RNA structure is thought to play a key role. To test this idea, we perform in-cellulo packaging experiments using bacteriophage MS2 coat proteins and a variety of RNA molecules in E. coli. In each experiment, plasmid-derived RNA molecules with a specified sequence compete against the cellular transcriptome for packaging by plasmid-derived coat proteins. Following this competition, we quantify the total amount and relative composition of the packaged RNA using electron microscopy, interferometric scattering microscopy, and high-throughput sequencing. By systematically varying the input RNA sequence and measuring changes in packaging outcomes, we are able to directly test competing models of selective packaging. Our results rule out a longstanding model in which selective packaging requires the well-known TR stem-loop, and instead support more recent models in which selectivity emerges from the collective interactions of multiple coat proteins and multiple stem-loops distributed across the RNA molecule. These findings establish a framework for understanding selective packaging in a range of natural viruses and virus-like particles, and lay the groundwork for engineering synthetic systems that package specific RNA cargoes.