Molecular regulation of retinal regeneration is context specific
Molecular regulation of retinal regeneration is context specific
Emmerich, K.; Hageter, J.; Hoang, T.; Lyu, P.; Sharrock, A. V.; Ceisel, A.; Thierer, J.; Chunawala, Z.; Nimmagadda, S.; Palazzo, I.; Matthews, F.; Zhang, L.; White, D. T.; Rodriguez, C.; Graziano, G.; Marcos, P.; May, A.; Mulligan, T.; Reibman, B.; Saxena, M. T.; Ackerley, D. F.; Qian, J.; Blackshaw, S.; Horstick, E.; Mumm, J. S.
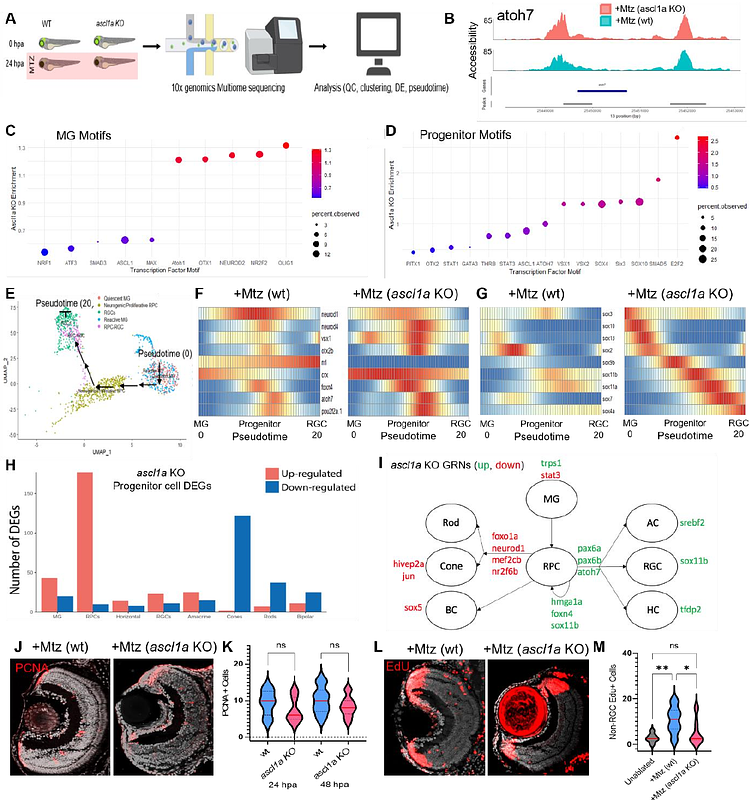
AbstractMany genes are known to regulate retinal regeneration following widespread tissue damage. Conversely, genes controlling regeneration following limited retinal cell loss, akin to disease conditions, are undefined. Combining a novel retinal ganglion cell (RGC) ablation-based glaucoma model, single cell omics, and rapid CRISPR/Cas9 based knockout methods to screen 100 genes, we identified 18 effectors of RGC regeneration kinetics. Surprisingly, 32 of 33 previously known/implicated regulators of retinal tissue regeneration were not required for RGC replacement; 7 knockouts accelerated regeneration, including sox2, olig2, and ascl1a. Mechanistic analyses revealed loss of ascl1a increased fate bias, the propensity of progenitors to produce RGCs. These data demonstrate plasticity and context-specificity in how genes function to control regeneration, insights that could help to advance disease tailored therapeutics for replacing lost retinal cells.