Backpropagation through time training of an unrolled Hodgkin Huxley model for automatic conductance estimation
Backpropagation through time training of an unrolled Hodgkin Huxley model for automatic conductance estimation
Li, Y.
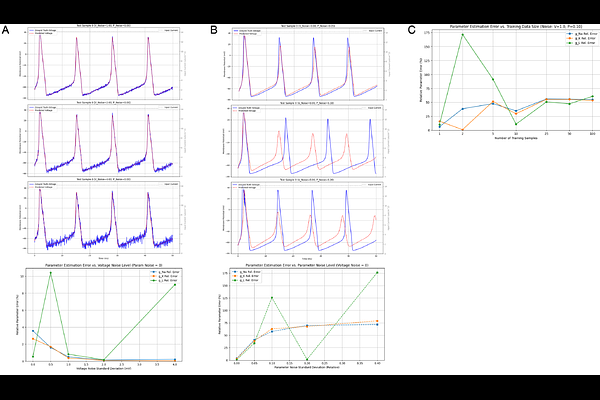
AbstractPrecise estimation of biophysical parameters such as ion channel conductances in single neurons is essential for understanding neuronal excitability and for building accurate computational models. However, inferring these parameters from experimental data is challenging, often requiring extensive voltage-clamp measurements or laborious manual tuning. Here we present a novel approach that leverages backpropagation through time (BPTT) to automatically fit a Hodgkin--Huxley (HH) conductance-based model to observed voltage traces. We unroll the HH model dynamics in time and treat the unknown maximum conductances as learnable parameters in a differentiable simulation. By optimizing the model to minimize the mean squared error between simulated and observed membrane voltage, our method directly recovers the underlying conductances (for sodium gNa, potassium gK, and leak gL) from a single voltage response. In simulations, the BPTT-trained model accurately identified conductance values across different neuron types and remained robust to typical levels of measurement noise. Even with a single current-clamp recording as training data, the approach achieved precise fits, highlighting its efficiency. This work demonstrates a powerful automated strategy for biophysical system identification, opening the door to rapid, high-fidelity neuron model customization from electrophysiological recordings. The code is availible at https://github.com/skysky2333/HH_BPTT.