Single Cell Analysis of Peripheral TB-Associated Granulomatous Lymphadenitis
Single Cell Analysis of Peripheral TB-Associated Granulomatous Lymphadenitis
Barrows, L.; Moos, P. J.; Carey, A. F.; Joseph, J.; Kialo, S.; Norrie, J.; Moyarelce, J. M.; Amof, A.; Nogua, H.; Lim, A. L.
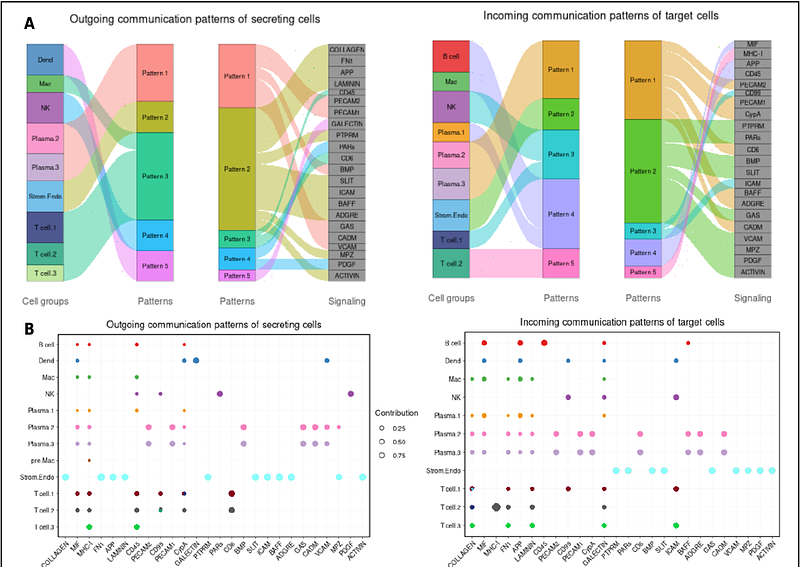
AbstractWe successfully employed a single cell RNA sequencing (scRNA-seq) approach to describe the cells and the communication networks characterizing granulomatous lymph nodes of TB patients. When mapping cells from individual patient samples, clustered based on their transcriptome similarities, we uniformly identify several cell types that characterize human and non-human primate granulomas. Whether high or low Mtb burden, we find the T cell cluster to be one of the most abundant. Many cells expressing T cell markers are clearly quantifiable within this CD3 expressing cluster. Other cell clusters that are uniformly detected, but that vary dramatically in abundance amongst the individual patient samples, are the B cell, plasma cell and macrophage/dendrocyte and NK cell clusters. When we combine all our scRNA-seq data from our current 23 patients (in order to add power to cell cluster identification in patient samples with fewer cells), we distinguish T, macrophage, dendrocyte and plasma cell subclusters, each with distinct signaling activities. The sizes of these subclusters also varies dramatically amongst the individual patients. In comparing FNA composition we noted trends in which T cell populations and macrophage/dendrocyte populations were negatively correlated with NK cell populations. In addition, we also discovered that the scRNA-seq pipeline, designed for quantification of human cell mRNA, also detects Mtb RNA transcripts and associates them with their host cells transcriptome, thus identifying individual infected cells. We hypothesize that the number of detected bacterial transcript reads provides a measure of Mtb burden, as does the number of Mtb-infected cells. The number of infected cells also varies dramatically in abundance amongst the patient samples. CellChat analysis identified predominating signaling pathways amongst the cells comprising the various granulomas, including many interactions between stromal or endothelial cells, such as Collagen, FN1 and Laminin, and the other component cells. In addition, other more selective communications pathways, including MIF, MHC-1, MHC-2, APP, CD 22, CD45, and others, are identified as originating or being received by individual immune cell components.


