CryoENsemble - a Bayesian approach for reweighting biomolecular structural ensembles using heterogeneous cryo-EM maps
CryoENsemble - a Bayesian approach for reweighting biomolecular structural ensembles using heterogeneous cryo-EM maps
Wlodarski, T.; Streit, J. O.; Mitropoulou, A.; Cabrita, L.; Vendruscolo, M.; Christodoulou, J.
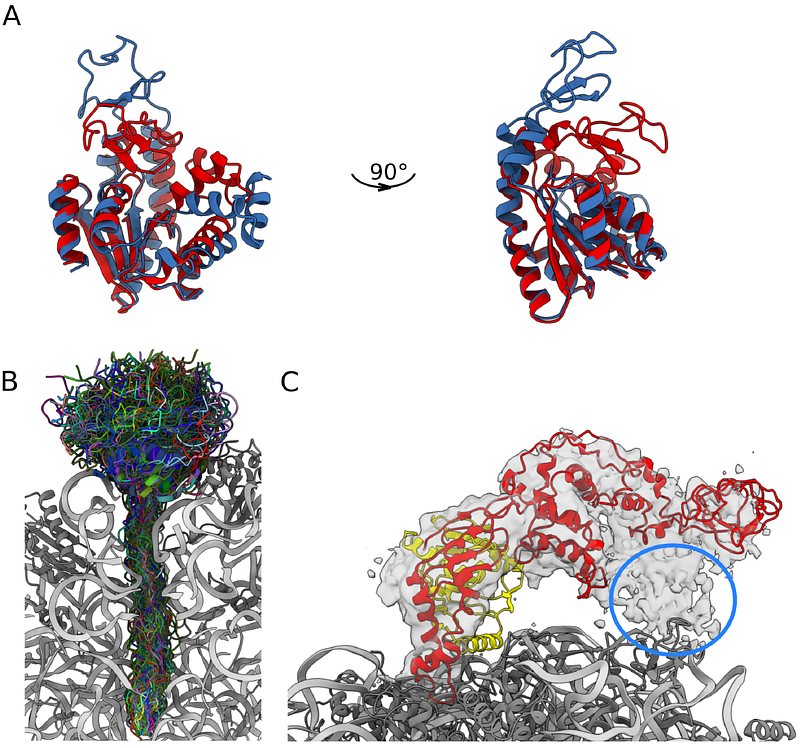
AbstractCryogenic electron microscopy (cryo-EM) has emerged as a central tool for the determination of structures of complex biological molecules. Accurately characterising the dynamics of such systems, however, remains a challenge. To address this, we introduce cryoENsemble, a method that applies Bayesian reweighing to conformational ensembles derived from molecular dynamics simulations to improve their agreement with cryo-EM data and extract dynamics information. We illustrate the use of cryoENsemble to determine the dynamics of the ribosome-bound state of the co-translational chaperone trigger factor (TF). We also show that cryoENsemble can assist with the interpretation of low-resolution, noisy or unaccounted regions of cryo-EM maps. Notably, we are able to link an unaccounted part of the cryo-EM map to the presence of another protein (methionine aminopeptidase, or MetAP), rather than to the dynamics of TF, and model its TF-bound state. Based on these results, cryoENsemble is expected to find use for challenging heterogeneous cryo-EM maps for various biomolecular systems, especially those encompassing dynamic elements.