Comparative genomics and transcriptomics on salt tolerance of Vigna luteola
Comparative genomics and transcriptomics on salt tolerance of Vigna luteola
Iki, Y.; Wang, F.; Ito, K.; Wakatake, T.; Tanoi, K.; Naito, K.
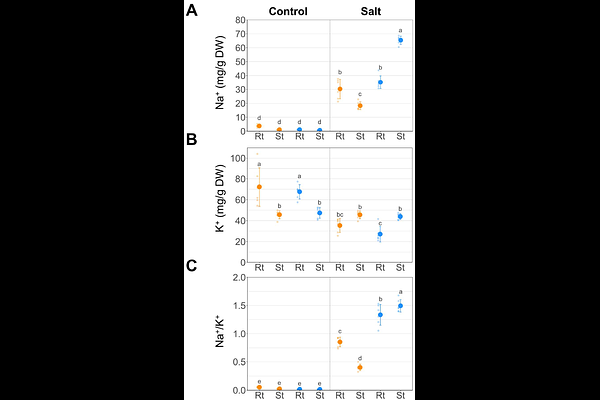
AbstractVigna luteola, a wild legume species, shows remarkable variation in salinity tolerance across its natural habitats, with coastal populations exhibiting high tolerance and riverbank populations being sensitive. This intraspecific variation provides a valuable system for investigating the genetic basis of salt tolerance. A major QTL for salt tolerance was previously identified by crossing salt-tolerant and salt-sensitive accessions, but the responsible genes remain unknown. In this study, grafting experiments between the two accessions revealed that the root plays a primary role in salt tolerance by suppressing Na transport to the shoot. We then conducted root transcriptome analysis and identified four candidate genes located within the QTL and highly expressed under salt stress in the tolerant accession: CBL-INTERACTING PROTEIN KINASE 6 (CIPK6), CAFFEOYL SHIKIMATE ESTERASE (CSE), FCS-LIKE ZINC FINGER PROTEIN 13 (FLZ13), and DROUGHT-INDUCED 21 (DI21). Promoter analysis revealed that the CIPK6 promoter contains transcription factor binding motifs unique to the salt-tolerant accession, which may contribute to its high expression under salt stress. These findings suggest that CIPK6 is regulated by cis-regulatory differences and is the most promising candidate for the salt-tolerance QTL. The identified genes in this study provide a foundation for developing salt-tolerant crops in the future.