Contrastive Learning for Omics-guided Whole-slide Visual Embedding Representation
Contrastive Learning for Omics-guided Whole-slide Visual Embedding Representation
Yu, S.; Kim, Y.; Kim, H.; Lee, S.; Kim, K.
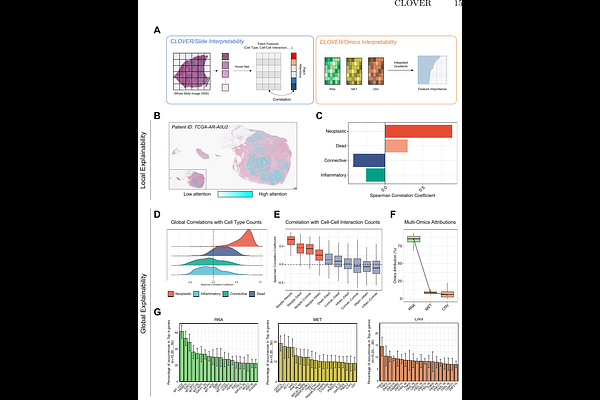
AbstractWhile computational pathology has transformed cancer diagnosis and prognosis prediction, existing computational methods remain limited in their ability to decipher the complex molecular characteristics within tumors. We present CLOVER (Contrastive Learning for Omics-guided whole-slide Visual Embedding Representation), a novel deep learning framework that leverages self-supervised contrastive learning to integrate multi-omics data (genomics, epigenomics, and transcriptomics) with slide representations, connecting the morphological and molecular features of tumors. Using the breast cancer cohorts comprising diagnostic slides and multi-omics paired data from 610 breast cancer patients, we validated CLOVER\'s excellence by demonstrating its ability to generate effective slide-level representations that consider molecular states of cancer. CLOVER outperforms existing methods in few-shot learning scenarios, particularly in cancer subtype classification and clinical biomarker prediction tasks (ER, PR, and HER2 status). Through comprehensive interpretability analysis, we identified tumor microenvironment components within slides and revealed molecular features associated with breast cancer. Our results demonstrate that CLOVER enables detailed molecular characterization from single slide analysis, suggesting its potential for effective utilization in future cancer diagnosis and prognosis prediction studies.