PyReconstruct: A fully opensource, collaborative successor to Reconstruct
PyReconstruct: A fully opensource, collaborative successor to Reconstruct
Chirillo, M. A.; Falco, J. N.; Musslewhite, M. D.; Lindsey, L. F.; Harris, K. M.
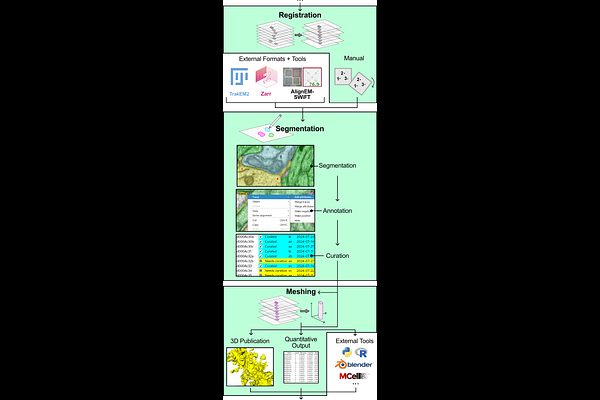
AbstractAs the serial section community transitions to volume electron microscopy, tools are needed to balance rapid segmentation efforts with documenting the fine detail of structures that support cell function. New annotation applications should be accessible to users and meet the needs of the neuroscience and connectomics communities while also being useful across other disciplines. Issues not currently addressed by a single, modern annotation application include: 1) built-in curation systems with utilities for expert intervention to provide quality assurance, 2) integrated alignment features that allow for image registration on-the-fly as image flaws are discovered during annotation, 3) simplicity for non-specialists within and beyond the neuroscience community, 5) a system to store experimental meta-data with annotation data in a way that researchers remain masked regarding condition to avoid potential biases, 6) local management of large datasets, 7) fully open-source codebase allowing development of new tools, and more. Here, we present PyReconstruct, a modern successor to the Reconstruct annotation tool. PyReconstruct operates in a field-agnostic manner, runs on all major operating systems, breaks through legacy RAM limitations, features an intuitive and collaborative curation system, and employs a flexible and dynamic approach to image registration. It can be used to analyze, display, and publish experimental or connectomics data. PyReconstruct is suited for generating ground truth to implement in automated segmentation, outcomes of which can be returned to PyReconstruct for proofreading and quality control.