ClOneHORT: Approaches for Improved Fidelity in Generative Models of Synthetic Genomes
ClOneHORT: Approaches for Improved Fidelity in Generative Models of Synthetic Genomes
Laboulaye, R.; Borda, V.; Chen, S.; North, K. E.; Kaplan, R.; O'Connor, T. D.
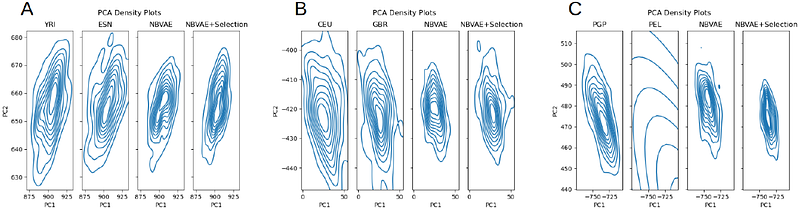
AbstractMotivation: Deep generative models have the potential to overcome difficulties in sharing individual-level genomic data by producing synthetic genomes that preserve the genomic associations specific to a cohort while not violating the privacy of any individual cohort member. However, there is significant room for improvement in the fidelity and usability of existing synthetic genome approaches. Results: We demonstrate that when combined with plentiful data and with population-specific selection criteria, deep generative models can produce synthetic genomes and cohorts that closely model the original populations. Our methods improve fidelity in the site-frequency spectra and linkage disequilibrium decay and yield synthetic genomes that can be substituted in downstream local ancestry inference analysis, recreating results with .91 to .94 accuracy.


