Diversification of gene expression across extremophytes and stress-sensitive species in the Brassicaceae
Diversification of gene expression across extremophytes and stress-sensitive species in the Brassicaceae
Wang, G.; Ryu, K. H.; Dinneny, A.; Carlson, J.; Goodstein, D. M.; Lee, J.; Oh, D.-H.; Oliva, M.; Lister, R.; Dinneny, J. R.; Schiefelbein, J.; Dassanayake, M.
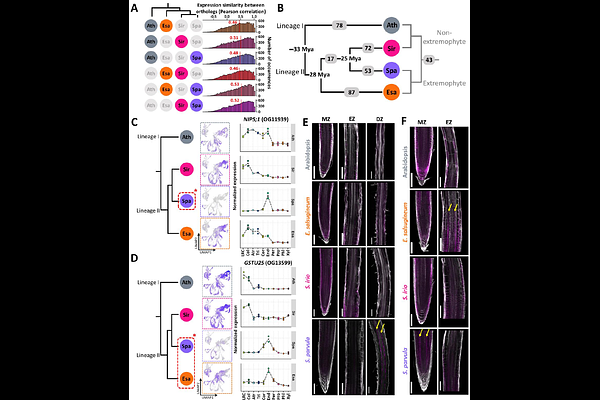
AbstractStress-sensitive and stress-adapted plants respond differently to environmental stresses. To explore the cellular-level stress adaptations, we built root single-cell transcriptome atlases for diverse Brassicaceae species: stress-sensitive plants (Arabidopsis thaliana and Sisymbrium irio), extremophytes (Eutrema salsugineum and Schrenkiella parvula) and a polyploid crop (Camelina sativa), under control, NaCl, and abscisic acid treatments. Approximately half of Arabidopsis cell-type markers lacked expression conservation across species. We identified new conserved cell-type markers, along with orthologs showing divergent expressions. We experimentally mapped distinct cortex sub-populations to different cortex layers across species. We found distinct cell-type-specific transcriptomic responses between species and treatments. Lineage-specific losses of stress responses were less prevalent but evolutionarily more favored than gains. In C. sativa, sub-genomes contributed equally to stress responses and homeologs with divergent stress responses typically do not exhibit high coding sequence or expression divergence. Our study provides a foundational root atlas and an analytical framework for multi-species single-cell transcriptomics.