Cenchrus purpureus and Cenchrus americanus repeatome provide chromosomal markers to distinguish subgenomes
Cenchrus purpureus and Cenchrus americanus repeatome provide chromosomal markers to distinguish subgenomes
Silvestrini, A. J. A.; Vaio, M.; Azevedo, K. C.; Torres, G. A.
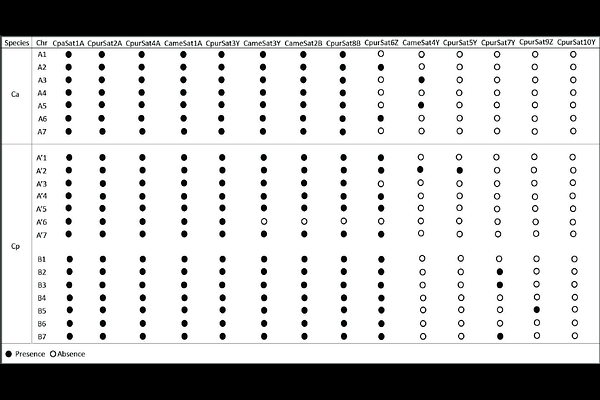
AbstractCenchrus L. is an important genus within the Poaceae family, comprising several species of high agronomic significance, such as Cenchrus purpureus and Cenchrus americanus, for production of forage and grains, respectively. Cenchrus americanus is a diploid species (2n = 2x = 14, AA genome), while Cenchrus purpureus is an allotetraploid (2n = 4x = 28, A1A1BB genome). The A1 subgenome is believed to be homeologous to and possibly derived from the A subgenome, while the origin of the B subgenome remains undefined. Despite their distinct subgenomic compositions, both species exhibit a high level of genome homology. The objective of the present work was the in silico characterization and comparative analysis of the repetitive fraction of the genomes of Cenchrus purpureus and Cenchrus americanus using genome skimming and a graph-based clustering approach, as well as the in situ hybridization of specific satellite DNA clusters into the chromosomes of both species. The repetitive fraction of the genome of C. purpureus and C. americanus corresponds to 52.23% and 76.82%, respectively. The most abundant repetitive elements in both species are the LTR retrotransposons. Satellite DNA sequences correspond to 2.55% and 4.17% of the genome of each species, respectively. Four new satellite sequences were identified as subgenome-specific sequences for both species, along with new centromeric variants. The ancestral relationship and the polyploidization-diploidization cycles played a fundamental role in the composition of their repetitive fraction. These cycles led Cenchrus americanus, a possible paleopolyploid, to a greater abundance of transposable elements compared to Cenchrus purpureus, a recent allopolyploid.