Close-Up of vesicular ER Exit Sites by Volume Electron Imaging using FIB-SEM
Close-Up of vesicular ER Exit Sites by Volume Electron Imaging using FIB-SEM
Nair, A.; Nair, A.; Stock, P.; Jain, A.; Somerville, E.; Sanyal, A.; Kirchhausen, T.
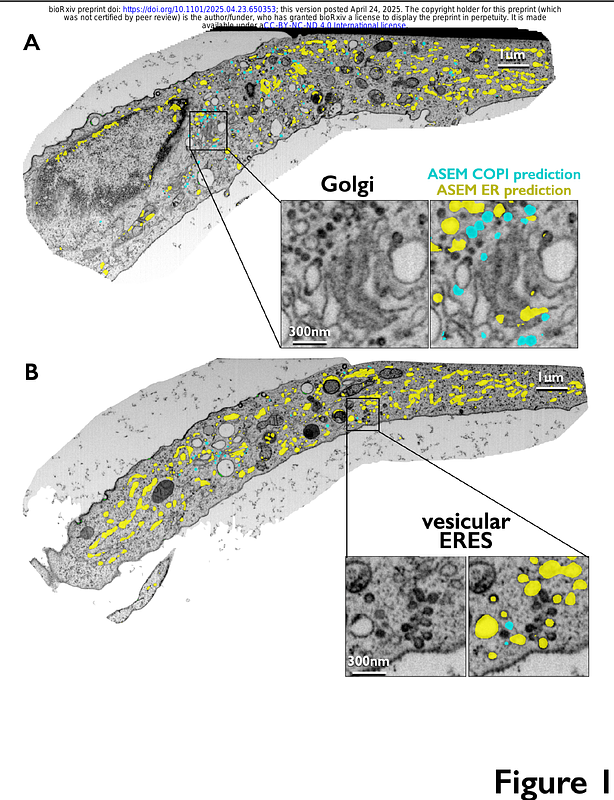
AbstractVolume electron microscopy by high-pressure freeze substitution combined with block-face focused ion beam scanning electron microscopy (FIB-SEM) provides comprehensive 3D views of subcellular architecture, essential for understanding cellular activity in context. Using Automated Segmentation in Electron Microscopy (ASEM)--a 3D-Unet convolutional neural network trained with sparse annotations--we characterized the spatial organization of endoplasmic reticulum exit sites (ERES), the initial locations for membrane remodeling in protein secretion, in cells not overexpressing secretory cargo. The ASEM model, trained on 50-70 nm in diameter COPI vesicles, successfully identified vesicles adjacent to the Golgi apparatus of HeLa, SVG-A, and iPSC-derived neurons. It also revealed abundant vesicles of similar appearance, often within a group of ~5-40 vesicles clustered within a 250 nm3 region adjacent to flattened endoplasmic reticulum (ER) domains, forming what we propose are COPII-mediated vesicular ER-exit sites. Elsewhere, smaller assemblies of 1-3 vesicles appeared alongside tubular ER networks emerging from similarly enlarged ER domains previously reported as the only ERES in HeLa cells. These findings underscore the power of large-scale volume electron microscopy to resolve contradictions regarding membrane organization in vesicular trafficking. We encourage the scientific community to use our publicly accessible repository, containing open-source code, trained models, annotations, predictions, and FIB-SEM datasets, to facilitate continued advances in automated segmentation methods.