Sequencing whole genomes of the West Javanese population in Indonesia reveals novel variants and improves imputation accuracy
Sequencing whole genomes of the West Javanese population in Indonesia reveals novel variants and improves imputation accuracy
Ardiansyah, E.; Riza, A.-L.; Dian, S.; Ganiem, A. R.; Alisjahbana, B.; Setiabudiawan, T. P.; Laarhoven, A. v.; Crevel, R. v.; Kumar, V.; ULTIMATE consortium,
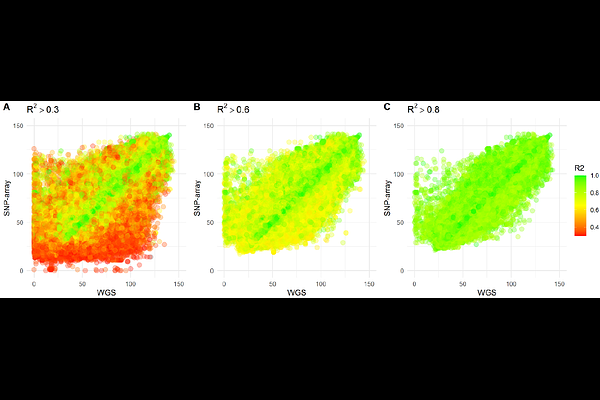
AbstractExisting genotype imputation reference panels are mainly derived from European populations, limiting their accuracy in non-European populations. To improve imputation accuracy for Indonesians, the fourth most populous country, we combined Whole Genome Sequencing (WGS) data from 227 West Javanese individuals with East Asian data from the 1000 Genomes Project. This created three reference panels: EAS 1KGP3 (EASp), Indonesian (INDp), and a combined panel (EASp+INDp). We also used ten West Javanese samples with WGS and SNP-typing data for benchmarking. We identified 1.8 million novel single nucleotide variants (SNVs) in the West Javanese population, which, while similar to the East Asians, are distinct from the Central Indonesian Flores population. Adding INDp to the EASp reference panel improved imputation accuracy (R2) from 0.85 to 0.90, and concordance from 87.88% to 91.13%. These findings underscore the importance of including Indonesian genetic data in reference panels, advocating for broader WGS of diverse Indonesian populations to enhance genomic studies.