Deep Volumetric Localization Super-Resolution Microscopy
Deep Volumetric Localization Super-Resolution Microscopy
Han, K.; Hua, X.; Qi, T.; Gao, Z.; Wang, X.; Jia, S.
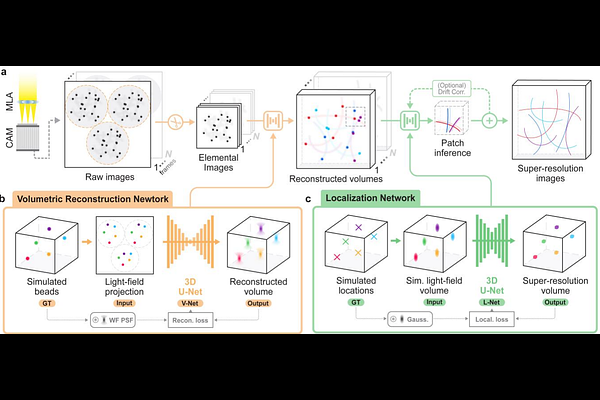
AbstractSuper-resolution microscopy, particularly localization-based methods, necessitates careful balancing of optical complexity, computational demands, and user accessibility. Conventional strategies typically adopt either deterministic or learning-based approaches, overlooking opportunities to leverage their synergistic strengths. In this work, we introduce deep volumetric localization microscopy (VLM), a super-resolution methodology that integrates instrumental and algorithmic advancements for high-fidelity 3D single-molecule imaging. VLM employs a wavefront-optimized light-field configuration to capture single-molecule data, while a cascaded neural network reconstructs 3D volumes and extracts molecular coordinates at a 10 nm and 25 nm localization precision in the lateral and axial dimensions, respectively, across an imaging depth exceeding 5 m. Unlike existing methods, VLM is trained exclusively with system-aware intrinsic point-spread functions, bypassing dependencies on external imaging modalities or sample-specific data training. We validate VLM across diverse biological specimens, demonstrating hardware simplicity, data efficiency, and minimal phototoxicity. We anticipate VLM will overcome current limitations in fluorescence microscopy, empowering broader advancements in biomedical research.