Single Cell Landscape of Sex-specific Drivers of Alzheimers Disease
Single Cell Landscape of Sex-specific Drivers of Alzheimers Disease
Wu, Y.; Travaglini, K. J.; Gabitto, M.; Keene, D.; Dunn, A.; Kaczorowski, C.; Harari, O.; De Jager, P.; Menon, V.; Schneider, J. A.; Bennett, D. A.; Dumitrescu, L.; Hohman, T. J.
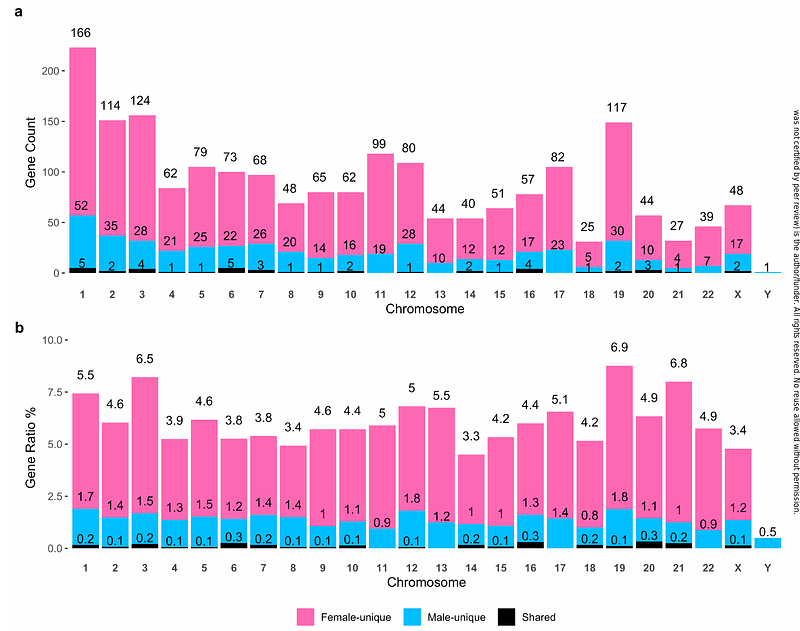
AbstractBackground Sex differences in Alzheimers disease (AD) have been documented for decades, and many sex-specific molecular contributors to AD have been discovered through bulk omics analysis of brain tissues. RNA sequencing (RNAseq) at single cell resolution provides an opportunity to characterize transcript associations with AD in a cell type-specific matter. Here, we investigated sex-specific gene expression associations with neuropathology and cognitive manifestation of AD (endophenotypes) leveraging a large single-nucleus transcriptomic dataset consisting of 1.64 million nuclei from dorsolateral prefrontal cortex (DLPFC) tissue of 424 unique donors from the Religious Orders Study and Memory and Aging Project (ROS/MAP; AD Knowledge Portal syn2580853). Methods ROS/MAP single-nucleus RNAseq data (snRNA-seq) were processed through a rigorous pipeline. In total, eight major cell types from DLPFC were identified from this dataset. We first performed sex-stratified and sex-interaction association analyses by fitting negative binomial mixed models in relation to {beta}-amyloid load (A{beta}), paired helical filament (PHF) tau tangle density (tau), global cognitive performance at last visit, and longitudinal cognitive trajectory. We then conducted gene set enrichment analysis to identify functional signaling pathways enriched for sex-specific associations. Lastly, we compared differential gene expression patterns and intercellular communication profiles between sexes and diagnostic groups among major cell types. For replication, sex-specific associations were examined using snRNA-seq derived from DLPFC tissue-derived of an independent set of 84 donors from The Seattle Alzheimers Disease Brain Cell Atlas (SEA-AD) study. Results 68% of the ROS/MAP participants were females, and 52% were diagnosed with AD dementia. We first two new disease vulnerable cell subpopulations including arterial smooth muscle cells (aSMC) within vascular niche and excitatory neuron subtype 3 (Exc.3). Then we identified 2,660 sex-specific associations involving 2,110 genes for A{beta} (51%), tau (21%), and cognitive performance (29%). Most (60%) female-specific associations were for A{beta}, and most (49%) male-specific associations were for tau. The vast majority (93%) of female protective associations were from neurons, and most (76%) of female risk associations were from glial cells. Nine of the female-specific associations involving eight unique genes were replicated from SEA-AD cohort, including ADGRV1 and OR3A3 with A{beta}, IFI27L1, LYRM1, STAP2, and TSTD2 with tau, PDYN with global cognition, and TMEM50B with longitudinal cognitive decline. All replicated associations except TMEM50B were observed in neurons. Furthermore, the preponderance of protective female-specific associations in neurons was also recapitulated in the SEA-AD cohort. Sex-specific gene associations were enriched for genes in the immune, inflammation, and damage-related stress response pathways, and microglia presented the most sex-specific enriched pathways. Finally, we identified six ITGB1-mediated microglia-specific incoming signals that may play a role in female-specific risk for {beta}-amyloid accumulation. Conclusion Our study highlights the transcriptome-wide single cell landscape of sex-specific molecular associations with AD neuropathology and cognitive decline. We provide a comprehensive atlas of sex-specific transcript associations, differential expression, signaling pathway, and cell-cell communication network changes in each major DLPFC cell type, while identifying and replicating several female-specific gene associations in neurons to help direct future mechanistic studies.