Single-cell SNP-DNA sequencing precisely maps genotoxic events in CRISPR-edited primary cells
Single-cell SNP-DNA sequencing precisely maps genotoxic events in CRISPR-edited primary cells
Boutin, J.; FAYET, S.; MARIN, V.; BERGES, C.; Riandiere, M.; Toutain, J.; Lamrissi-Garcia, I.; THIBAULT, C.; Cappellen, d.; Dabernat, S.; Poulet, A.; Francilette, M.; DROUIN, N.; MOREAU-GAUDRY, F.; BEDEL, A.
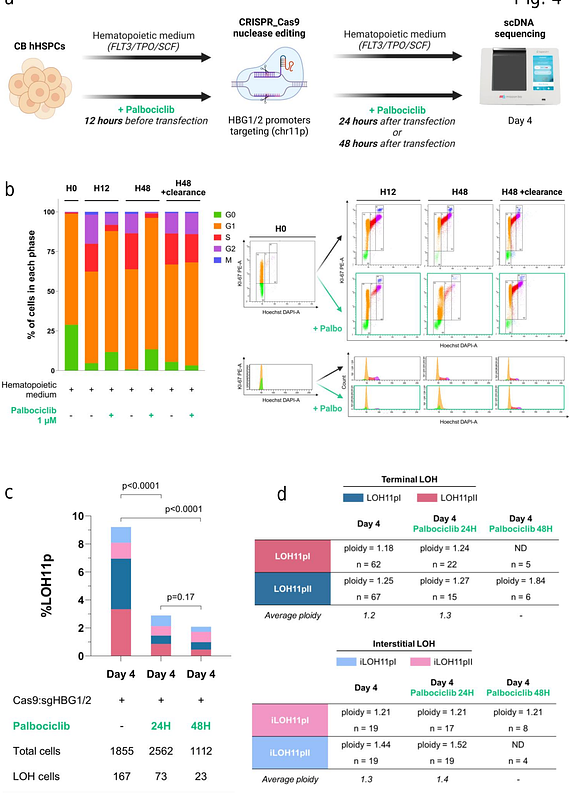
AbstractGenome editing by CRISPR-Cas9 is promising for genetic disease and cancer gene therapy. However, safety concerns are still present, particularly the ON-target genotoxicity for protocols using nucleases. Quality control of edited cells before and after graft is mandatory, especially to assay megabase-scale genomic rearrangements induced at the targeted locus. These unintended events are fortunately rare but potentially deleterious. Classical PCR-based bulk approaches do not detect them or underestimate their frequency. Single-cell approaches are promising but RNA sequencing only estimates copy number variation of the genome. Here, we propose single-cell DNA sequencing to accurately evaluate and monitor CRISPR-mediated genotoxicity in primary cells (human fibroblasts and hematopoietic stem/progenitor cells). We designed a homemade panel using single nucleotide polymorphisms to detect, map and characterize induced-losses of heterozygosity (terminal, interstitial, copy-loss and copy-neutral). This innovative approach revealed intense genotoxicity linked to the double strand break. The risk was associated with DNA repair modulators in particular DNA-PKcs inhibitor AZD7648. Importantly, the CDK4/6 inhibitor palbociclib prevented these rearrangements in hematopoietic stem/progenitor cells. This work strongly suggests that single-cell DNA sequencing should be routinely implemented in clinical applications before CRISPR-edited cell infusions.