Decoding the Centromeric Region with a Near Complete Genome Assembly of the Oshima Cherry Cerasus speciosa
Decoding the Centromeric Region with a Near Complete Genome Assembly of the Oshima Cherry Cerasus speciosa
Fujiwara, K.; Toyoda, A.; Biswa, B. B.; Kishida, T.; Tsuruta, M.; Nakamura, Y.; Kimura, N.; Kawamoto, S.; Sato, Y.; Katsuki, T.; Sakura 100 Genome Consortium, ; Koide, T.
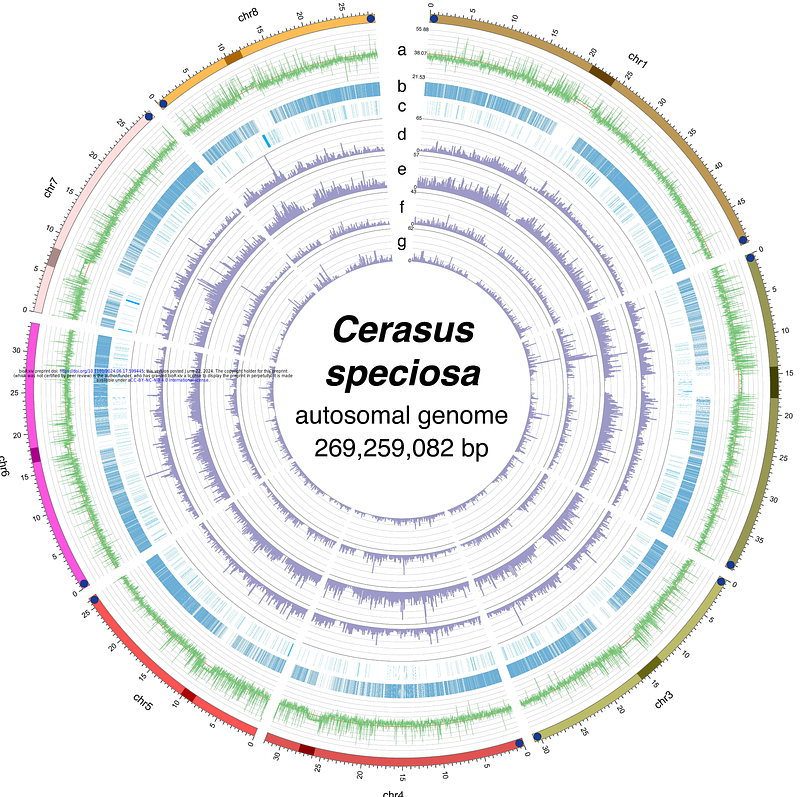
AbstractThe Oshima cherry (Cerasus speciosa), which is endemic to Japan, has significant cultural and horticultural value. In this study, we present a near complete telomere-to-telomere genome assembly for C. speciosa, derived from the old growth \"Sakurakkabu\" tree on Izu Oshima Island. Using Illumina short-read, PacBio long-read, and Hi-C sequencing, we constructed a 269.3 Mbp genome assembly with a contig N50 of 32.0 Mbp. We examined the distribution of repetitive sequences in the assembled genome and identified regions that appeared to be centromeric. Detailed structural analysis of these putative centromeric regions revealed that the centromeric regions of C. speciosa comprised repetitive sequences with monomer lengths of 166 or 167 bp. Comparative genomic analysis with Prunus sensu lato genome revealed structural variations and conserved syntenic regions. This high-quality reference genome provides a crucial tool for studying the genetic diversity and evolutionary history of Cerasus species, facilitating advancements in horticultural research and the preservation of this iconic species.