GeneGPT: Augmenting Large Language Models with Domain Tools for Improved Access to Biomedical Information
GeneGPT: Teaching Large Language Models to Use NCBI Web APIs
Qiao Jin, Yifan Yang, Qingyu Chen, Zhiyong Lu
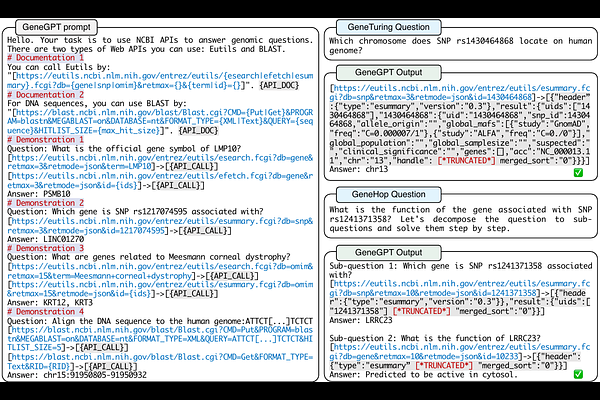
AbstractIn this paper, we present GeneGPT, a novel method for teaching large language models (LLMs) to use the Web Application Programming Interfaces (APIs) of the National Center for Biotechnology Information (NCBI) and answer genomics questions. Specifically, we prompt Codex (code-davinci-002) to solve the GeneTuring tests with few-shot URL requests of NCBI API calls as demonstrations for in-context learning. During inference, we stop the decoding once a call request is detected and make the API call with the generated URL. We then append the raw execution results returned by NCBI APIs to the generated texts and continue the generation until the answer is found or another API call is detected. Our preliminary results show that GeneGPT achieves state-of-the-art results on three out of four one-shot tasks and four out of five zero-shot tasks in the GeneTuring dataset. Overall, GeneGPT achieves a macro-average score of 0.76, which is much higher than retrieval-augmented LLMs such as the New Bing (0.44), biomedical LLMs such as BioMedLM (0.08) and BioGPT (0.04), as well as other LLMs such as GPT-3 (0.16) and ChatGPT (0.12).