KidneyTalk-open: No-code Deployment of a Private Large Language Model with Medical Documentation-Enhanced Knowledge Database for Kidney Disease
KidneyTalk-open: No-code Deployment of a Private Large Language Model with Medical Documentation-Enhanced Knowledge Database for Kidney Disease
Yongchao Long Department of Computer Science, Tianjin University of Technology, Tianjin, China National Institute of Health Data Science, Peking University, Beijing, China, Chao Yang Renal Division, Department of Medicine, Peking University First Hospital, Beijing, China Center for Digital Health and Artificial Intelligence, Peking University First Hospital, Beijing, China, Gongzheng Tang National Institute of Health Data Science, Peking University, Beijing, China, Jinwei Wang Renal Division, Department of Medicine, Peking University First Hospital, Beijing, China, Zhun Sui Renal Department, Peking University People's Hospital, Beijing, China, Yuxi Zhou Department of Computer Science, Tianjin University of Technology, Tianjin, China Institute of Internet Industry, Tsinghua University, Beijing, China, Shenda Hong National Institute of Health Data Science, Peking University, Beijing, China Department of Emergency Medicine, Peking University First Hospital, Beijing, China, Luxia Zhang National Institute of Health Data Science, Peking University, Beijing, China Renal Division, Department of Medicine, Peking University First Hospital, Beijing, China Research Units of Diagnosis and Treatment of Immune-Mediated Kidney Diseases, Chinese Academy of Medical Sciences, Beijing, China State Key Laboratory of Vascular Homeostasis and Remodeling, Peking University, Beijing, China Center for Digital Health and Artificial Intelligence, Peking University First Hospital, Beijing, China
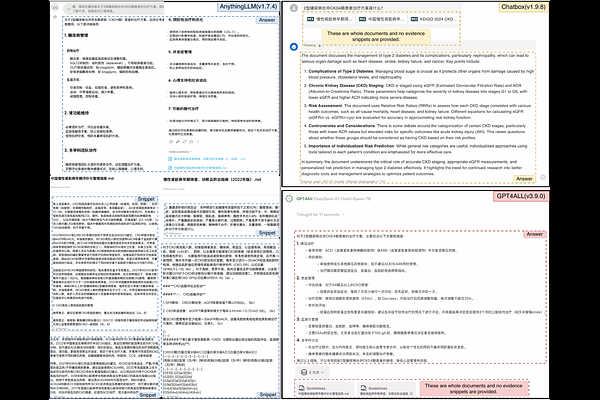
AbstractPrivacy-preserving medical decision support for kidney disease requires localized deployment of large language models (LLMs) while maintaining clinical reasoning capabilities. Current solutions face three challenges: 1) Cloud-based LLMs pose data security risks; 2) Local model deployment demands technical expertise; 3) General LLMs lack mechanisms to integrate medical knowledge. Retrieval-augmented systems also struggle with medical document processing and clinical usability. We developed KidneyTalk-open, a desktop system integrating three technical components: 1) No-code deployment of state-of-the-art (SOTA) open-source LLMs (such as DeepSeek-r1, Qwen2.5) via local inference engine; 2) Medical document processing pipeline combining context-aware chunking and intelligent filtering; 3) Adaptive Retrieval and Augmentation Pipeline (AddRep) employing agents collaboration for improving the recall rate of medical documents. A graphical interface was designed to enable clinicians to manage medical documents and conduct AI-powered consultations without technical expertise. Experimental validation on 1,455 challenging nephrology exam questions demonstrates AddRep's effectiveness: achieving 29.1% accuracy (+8.1% over baseline) with intelligent knowledge integration, while maintaining robustness through 4.9% rejection rate to suppress hallucinations. Comparative case studies with the mainstream products (AnythingLLM, Chatbox, GPT4ALL) demonstrate KidneyTalk-open's superior performance in real clinical query. KidneyTalk-open represents the first no-code medical LLM system enabling secure documentation-enhanced medical Q&A on desktop. Its designs establishes a new framework for privacy-sensitive clinical AI applications. The system significantly lowers technical barriers while improving evidence traceability, enabling more medical staff or patients to use SOTA open-source LLMs conveniently.